The Brain’s Stunning Genomic Diversity Revealed
(Originally published by the Salk Institute)
September 9, 2016
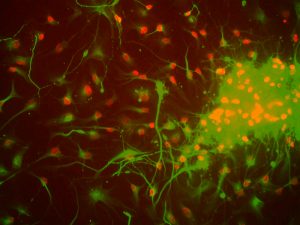
Our brains contain a surprising diversity of DNA. Even though we are taught that every cell in our body has the same DNA, in fact most cells in the brain have changes to their DNA that make each neuron a little different.
Now researchers at the Salk Institute and their collaborators have shown that one source of this variation—called long interspersed nuclear elements or L1s—are present in 44 to 63 percent of healthy neurons and can not only insert DNA but also remove it. Previously, these L1s were known to be small bits of DNA called “jumping genes” that copy and paste themselves throughout the genome, but the researchers found that they also cause large deletions of entire genes. What’s more, such variations can influence the expression of genes that are crucial for the developing brain.
The findings, published September 12, 2016 in the journal Nature Neuroscience, may help explain what makes us each unique—why even identical twins can be so different from one other, for example—and how jumping genes can go awry and cause disease.
“In 2013, we discovered that different neurons within the same brain have various complements of DNA, suggesting that they function slightly differently from each other even within the same person,” says the study’s senior investigator Rusty Gage, a professor in Salk’s Laboratory of Genetics and holder of the Vi and John Adler Chair for Research on Age-Related Neurodegenerative Diseases; Gage is also a co-director of the Kavli Institute for Brain and Mind at UC San Diego. “This recent study reveals a new and surprising form of variation that will help us understand the role of L1s, not only in healthy brains but in those affected by schizophrenia and autism.”
In 2005, Gage’s team discovered L1s as a mechanism of genome diversity in the brain. However, it was not until it became possible to sequence the entire genome of a single cell that scientists could get a handle on the amount and nature of these variations. Using single-cell sequencing detailed in a 2013 Science paper, Gage’s group showed that large chunks of DNA were inserted—or deleted—into the genomes of the cells.
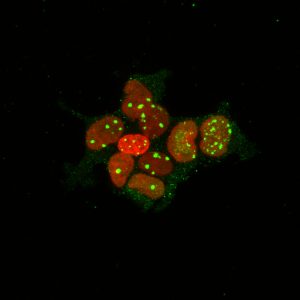
But even in that study the mechanisms responsible for causing insertions and deletions were unclear, making it difficult to decipher whether specific regions of the genome were more or less likely to be altered, as well as whether jumping genes were related to the deletions.
In the new study, Gage, co-first authors Jennifer Erwin and Apuã Paquola, and collaborators developed a method to better capture the L1-associated variants in healthy neurons for sequencing and created a computational algorithm to distinguish the variations with greater accuracy than before.
Using stem cells that are coaxed to differentiate into neurons in a dish, the team found that L1s are prone to DNA breaks. That’s because a specific enzyme that chews through L1 spots in the genome is particularly active during differentiation. People inherit some L1s from their parents, and the enzyme appears to cut near these spots, the group found.
“The surprising part was that we thought all L1s could do was insert into new places. But the fact that they’re causing deletions means that they’re affecting the genome in a more significant way,” says Erwin, a staff scientist in Gage’s group.
Gage believes that diversity can be good for the brain—after all, about half of our brain cells have large chunks of missing or inserted DNA caused by L1s alone—but that too much of it can cause disease.
Recent evidence has shown that neurons derived from individuals with schizophrenia or the rare autism-associated disorder Rett syndrome harbor more than normal amounts of L1 variations in their genomes. In the new study, the team examined a schizophrenia-associated gene called DLG2, in which introducing L1 variations can change the gene’s expression and subsequent maturation of neurons. The group plans to explore the role of L1 variations in other genes and their effects on brain activity and disease.

Other authors on the study are Tatjana Singer, Iryna Gallina, Carolina Quayle, Tracy Bedrosian, Cheyenne Butcher, Joseph Herdy and Anindita Sarkar of Salk; Mark Novotny and Roger Lasken of the J. Craig Venter Institute; Francisco Alves of the University of São Paulo in Brazil; and Alysson Muotri of the University of California, San Diego.
The research was supported by George E. Hewitt Foundation for Medical Research, the California Institute for Regenerative Medicine, the National Institutes of Health (MH095741, MH088485), the G. Harold & Leila Y. Mathers Foundation, the Engman Foundation, the Leona M. and Harry B. Helmsley Charitable Trust, the Paul G. Allen Family Foundation, the Glenn Center for Aging Research at the Salk Institute, and JPB Foundation.