From Fins to Limbs: New Research Offers Clues to How Animals Left the Sea and Evolved to Walk on Land
(Originally published by the Zuckerman Mind Brain Behavior Institute)
January 11, 2018
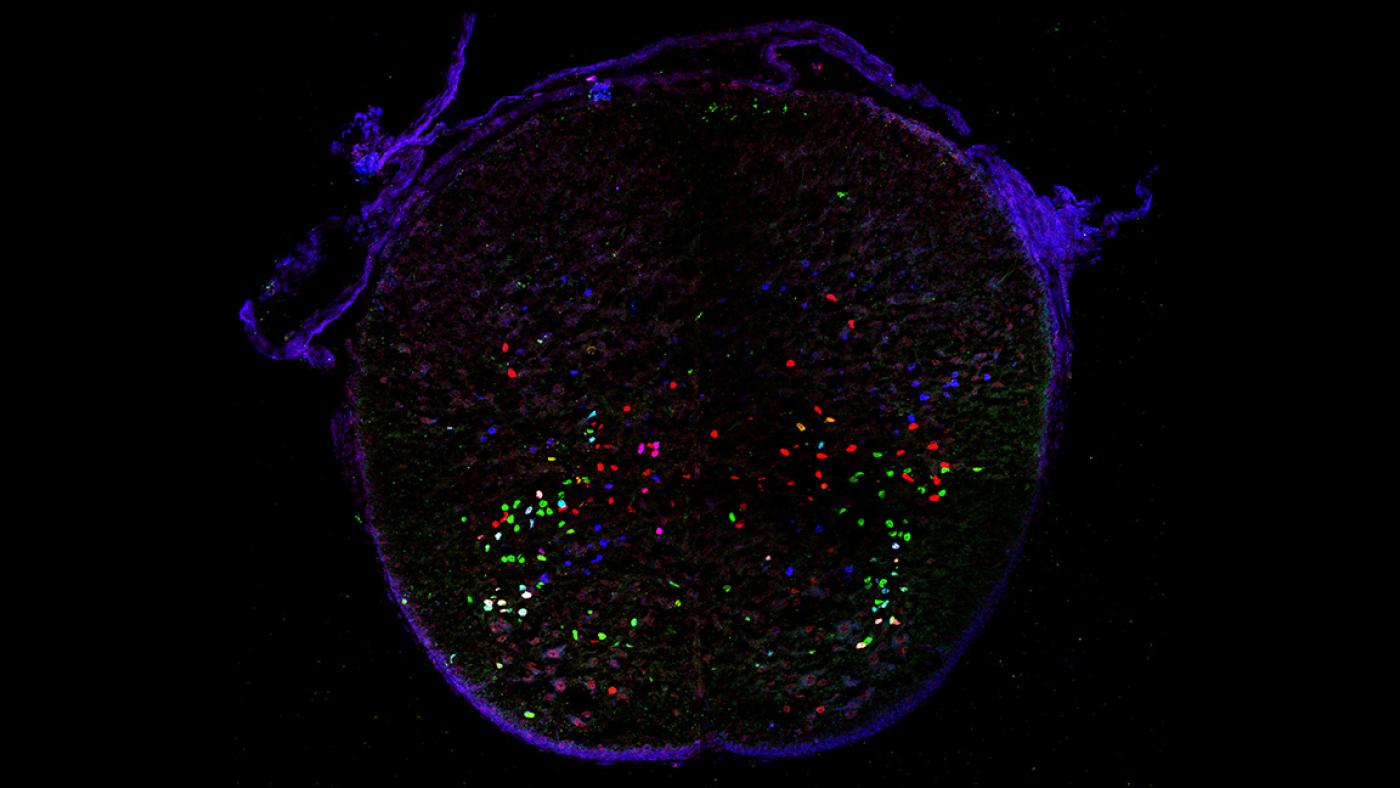
Partially overlapping expression of three transcription factors in from thoracic region of mouse spinal cord (Credit: Lora Sweeney/Jessell and Kintner Labs/Salk Institute)
Four-hundred million years ago, when the Earth was surrounded by a single, vast ocean, when North America and Europe shared a common border, a strange, fish-like creature performed an extraordinary feat: it took a step.
The animal likely resembled a fish, amphibian and reptile all rolled into one and it moved differently than its ancestors. Instead of gliding through the water, it could walk, and likely split its time between sea and land. Its descendants evolved to become even more adept at walking and ventured farther afield. Their adaptable limbs (which sometimes became wings) propelled them to nearly every inhabitable region on the planet.
This transition from sea to land marks one of biology’s most important accomplishments. The fossil record has revealed a wealth of information about this point in time, but many questions remain. In recent years, scientists have looked to clues hidden in living tissue, examining the cells that together comprise the spinal cord and therefore guide every aspect of movement.
In new research published in Neuron, scientists at Columbia University’s Mortimer B. Zuckerman Mind Brain Behavior Institute and the Salk Institute classified differences between some of these cells in mice, and in so doing opened a window into the evolutionary history of this life-altering behavior. We spoke with the paper’s senior author Thomas M. Jessell, PhD, codirector of the Zuckerman Institute as well as Columbia's Kavli Institute for Brain Science, and first author Lora Sweeney, PhD, a postdoctoral fellow at the Zuckerman and Salk Institute jointly in the labs of Dr. Jessell’s and the Salk Institute’s Christopher Kintner, PhD, about this research.
What is the big question you wanted to answer?
Thomas M. Jessell: Our particular interest lies in the evolutionary origins of limbs themselves. More precisely, we want to know how nerve cells called inhibitory interneurons, one of many cell types that regulate muscle movement, evolved and arranged themselves in such a way as to facilitate walking on land.
For example, the way a fish moves in the water is vastly different from how an animal moves on land. A fish’s undulatory, swimming motion is largely controlled by the muscles that run along the middle of its torso, called thoracic muscles. These muscles are ancient, and variations of them still exist in all land animals alive today — including humans.
By contrast, limbs are controlled by different sets of muscles. The movements that limbs can produce — walking, grasping, reaching — are specific to land dwellers.
Lora Sweeney: Because different muscles performed different actions, it stood to reason that the interneurons that controlled different muscle types would be distinct as well. In fact, decades of research across the field argued if differences in these interneurons existed, we should be able to find them and classify them, thus creating a map of movement across the entire spinal cord.
How could these differences be teased apart?
LS: Every neuron in the body contains the same complete set of genetic information. But what makes one neuron different from the next is which of those genes is switched on, and which is switched off, and in what order. Each neuron’s permutation of gene activity serves as its ‘identity’ — like a genetic fingerprint.
Last year, fellow members of the Jessell Lab, Drs. Jay Bikoff and Mariano Gabitto, devised an ingenious method to collect and analyze the genetic activity of interneurons in one spinal cord region. They then developed a robust computer algorithm that distinguished different cell types based on their genetic identities.
For our latest study, we took the principles established by Drs. Bikoff and Gabitto and applied them to a broader swath of interneurons in the spinal cords of mice. First, we examined interneurons from the thoracic, or torso, region, which regulate movement of the back and abdominal muscles. We then examined interneurons known to regulate the movement of the animals’ hind and forelimbs, which are used for walking but also for grasping and reaching.
What did you find?
LS: We hypothesized a couple of different outcomes. For example, interneurons controlling the thoracic and limb regions could be wholly intermingled with each other up and down the spinal cord, like a complicated spider web of cellular circuits.
In actuality, our findings revealed the opposite. Interneurons that controlled the thoracic region were clustered together into groups, while those that controlled limbs were clustered separately. But there was a strong element of layering to this separation. The thoracic interneurons appeared to have been laid down first, with the limb interneurons overlaid on top.
These observations evoked an evolutionary component to its organization. The older, more primitive thoracic region (that is present in both fish and land animals) evolved first. The more newly evolved neurons that controlled limb movement, which are only present in land animals, were added later.
Though these findings were in living cells, taken together they provided a window into the 400-million-year history of limb movement; and can serve as first glimpse into how the ability to walk may have arisen.
We are working to better understand the sea-to-land transition. Moving forward, we are beginning to examine the organization of inhibitory interneurons in an animal that exhibits both evolutionary states over the course of a single lifetime: the frog. We are currently studying the organization of the frog’s motor and interneurons at various life stages, from tadpole to adult, to see how neuron types may grow, move or otherwise change over time.
TMJ: This work also uncovered new insight into Hox genes, a versatile family of genes that helps to organize the body during development. Previous research had shown that Hox genes guide the placement of motor neurons (another type of nerve cell involved in movement). Our new research revealed that Hox genes are also largely responsible for the organization and placement of interneurons. Indeed, it appears that Hox genes play an even more important role than previously thought. By simultaneously coordinating the development and organization of different, but interconnected, neuron types, they likely act as a master regulator — ensuring every cell ends up in the right place and develops into integrated, functioning neural circuits.
Do your findings have clinical applications or help to inform disorders of movement, such as ALS?
LS: Amyotrophic lateral sclerosis, also known as Lou Gehrig’s disease, is essentially the progressive death of neurons responsible for movement, which causes an inability to move and ultimately results in death.
We know that different types of neurons, including inhibitory interneurons, are impacted by ALS in different ways. For example, neurons that control the limbs are particularly susceptible. But neurons in the thoracic region are some of the most resistant and are among the last to succumb to cell death.
TMJ: Figuring out why some cell types are resistant is key to understanding why other cell types are so susceptible. Our latest findings offer new insight into key differences in neurons of the thoracic region, and could be harnessed to better understand why they are more resistant to the progression of ALS.
Is it their differing evolutionary histories or molecular identities? Or perhaps differing ways in which these neurons organize themselves? These are all open questions that we will continue to investigate as we drive this research forward.
###
This paper is titled: “Origin and segmental diversity of spinal inhibitory interneurons.” Additional contributors include co-second authors Jay Bikoff, PhD and Mariano Gabitto, PhD, Susan Brenner-Morton, Myungin Baek, Jerry Yang, Esteban Tabak, Jeremy Dasen, PhD, and Christopher Kintner, PhD.
This research was supported by the Waitt Advanced Biophotonics Core Facility of the Salk Institute, by the National Cancer Institute (P30 014195), the National Institute of Neurological Disease and Stroke (NS072031), the Waitt Foundation, the Damon Runyun Cancer Foundation, Project ALS, the Brain Research Foundation, the Office of Naval Research, the National Science Foundation, the Harold and Leila Y. Mathers Foundation, the National Institutes of Health (R01 NS062822, R01NS097550, R01 GM076507, R01 NS033245) and the Howard Hughes Medical Institute.