From Nanoparticles to Nano Knots
by Alan S. Brown
Research Highlights from Kavli Nanoscience Institutes
The Author
At Kavli Nanoscience Institutes, researchers push the boundaries of our knowledge and capabilities of science at the nanoscale.
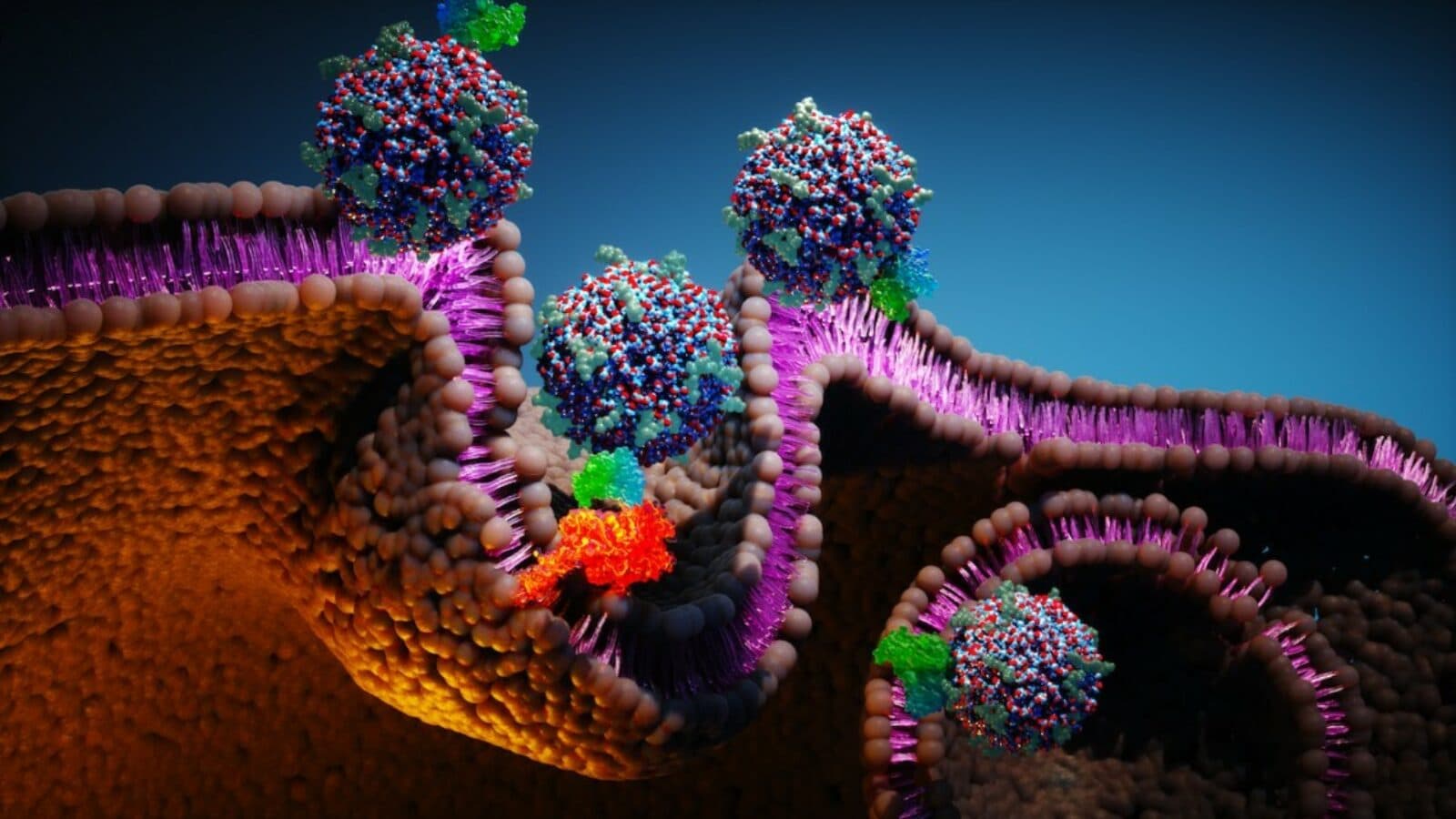
Nanoparticles with antibody fragments eradicate gastric cancer in mice.
When Ulrich Wiesner, a member of the Kavli Institute at Cornell, began developing silica-encased fluorescent nanoparticles 20 years ago, he hoped to use them in displays and sensors. Then he teamed with colleague Michelle Bradbury at Memorial Sloan Kettering and Weill Cornell Medicine to turn these nanoparticles, now called ‘C dots,’ into cancer killers. Working with AstraZeneca, the researchers loaded the 7-nanometer-sized C dots with antibody fragments, which target specific cancer tissue types, and five times the medication carried by most antibody medicines. Unlike antibody medications, which can accumulate and cause toxic reactions, C dots either kill tumor cells or get cleared from the body through the kidneys without accumulation. Three doses of C dot medicine eradicated the cancer in every mouse tested, with no evidence of tumor recurrence after nearly 200 days (mice live about 730 days). In 2014, the researchers formed a company, Elucida Oncology, that has raised $72 million to develop the technology.
Could polymers replace natural enzymes?
Could biology work just as well by substituting simpler polymers for natural enzymes? That is the intriguing question asked by Ting Xu, a member of Kavli ENSI at Berkeley. Instead of creating large molecules from 20 amino acids, she is using just a handful of chemical building blocks to create polymers that perform some of the same functions as natural proteins. The payoff: Xu’s proteins are much easier to synthesize than tweaking natural proteins and could be used to protect stents and other implants from attack by the body’s immune system, to make coatings that deliver drugs, and create biocompatible products like artificial tears and cartilage.
Cell-moving bubbles dance to a happy beat.
Nanoscale biology often involves moving individual cells, but how do you do it? One way is to use optical tweezers, originally developed to manipulate single molecules and structures within cells, but the high intensity light needed to move an entire cell could damage the specimen. Mikhail Shapiro, a member of the Kavli Nanoscience Institute at Caltech, has another approach. For several years, he has used gas vesicles, hollow protein shells that entrap air, to scatter sound so he can use ultrasound to image the location of bacterial cells as well as gene expression in the body. This led to Di Wu, a research scientist in his lab, to develop a technique to encapsulate one or more cells in a vesicle and use ultrasound to manipulate pressure and push the vesicle around. Potential uses from guiding cell-based therapies to a specific location or creating tissues layered with different types of cells to better mimic natural tissue for research purposes.
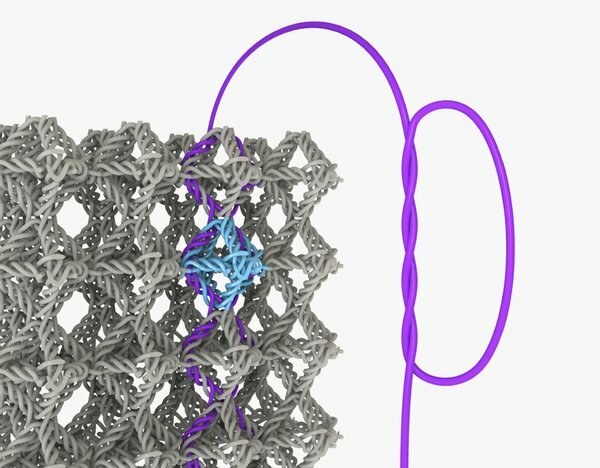
Nano knots are not for naught.
Julia Greer, director of the Kavli Nanoscience Institute at Caltech, is well know for creating nanoscale architected structures ranging from multiscale lattices to weaves to see how they affect material properties. Now, a Ph.D. student in her lab, Widianto Moestopo, has published a paper showing how materials made with nanoscale knots make materials far tougher than their unknotted counterparts. This is unusual, because usually nanoscale reinforcements make materials stronger but more brittle. The researchers made the knotted materials from polymers using Greer’s advanced 3D lithography system. When deformed, they bounced back to their original shape. Compared with their unknotted cousins, the knotted materials withstood more than twice the strain before snapping. Such materials could prove useful in aerospace structures and perhaps even medical sutures. More importantly, they are outstanding model systems for understanding friction and energy dissipation in nanomaterials.
2D Kondo sandwich creates ‘spherical cow’ for quantum researchers.
Sixty years ago, Jun Kondo explained how random magnetic impurities in a material could change its resistivity. Yet the quantum behavior of a regular lattice of impurities remained a mystery. Studying this phenomena usually involves intermetallic compounds of rare earth elements and trillions of interacting electrons. That’s enough to overwhelm theorists, who instead create what they refer to as a “spherical cow” Kondo lattice model to study this behavior. But two Kavli Institute at Cornell researchers, Drs. Kin Fai Mak and Jie Shan, have created a system to study these materials more controllably. By twisting atomically thin (2D) layers of molybdenum ditelluride and tungsten diselenide 180 degrees, electrons can be trapped like eggs in a carton – and properties like spin density and mass can be modified with voltage. The model system gives theorists a way to check their predictions and simplifies the study of heavy fermions, exotic quantum phase transitions, and other quantum behavior.