A Living Cell, Built from the Ground Up
by Alan S. Brown
The ultimate building challenge in biology
The Author
The Researcher
When Christophe Danelon was interviewing for a position at Delft University of Technology 10 years ago, someone asked him how long it would take to build a living synthetic cell.
“I said that 10 years seemed reasonable,” Danelon replied.
Now, after 10 years at Delft’s Kavli Institute of Nanoscience’s Department of Bionanoscience, Danelon admits that he was naive.
“But I think you need that naivete to embark on such a project,” he added. “There are some very challenging aspects of this work, and I can almost guarantee that in another 10 years we will not tackle all of them.”
Clearly, unraveling and then recreating the inner mechanisms of a single-celled organism, like a bacterium, is a monumental challenge. Yet researchers have made giant strides over the past few years. Danelon’s pioneering work in recreating the initial steps in cell division in an artificial cell is one of those breakthroughs.

The work was done under BaSyC (shorthand for "Building a Synthetic Cell"), The Netherlands’s 10-year research program to build a synthetic cell from the ground up. Scientists hope this will teach them to create simple organisms to do things like attack cancer, synthesize drugs and chemicals, and clean the environment.
Danelon, however, is more interested in unraveling the fundamental design principles of life. He calls it “the ultimate building challenge in biology.”
Building a cell as complicated and robust as even the simplest bacteria may take decades, he said. But a scaled-down version of that bacteria—a synthetic cell that could sense its environment, grow, and divide—could be possible within five years, he believes.
Lipids
As a young child, Danelon often wondered what made things alive. Yet as he grew older, his studies gravitated towards physics, chemistry, and math. It was only while finishing his master’s degree studies that he became interested in biology. His goal, when pursuing his Ph.D. and post-doctoral studies, was to use physics to understand the mechanisms of life.
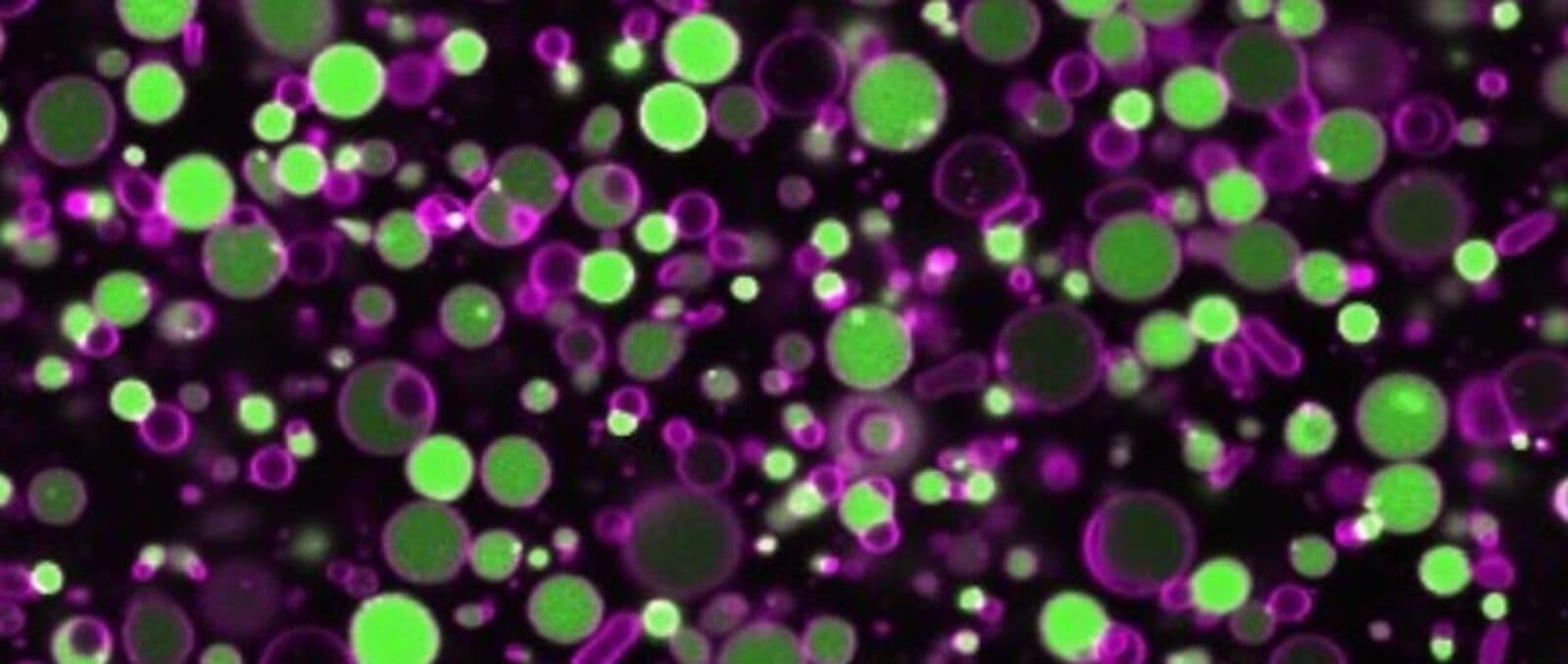
Along the way, be became fascinated by vesicles, microscopic containers that assemble spontaneously from lipids—long fatty acid molecules like the ones that make up the membranes of cells.
Like cells, lipid vesicles can grow, shrink, and deform due to external conditions, Danelon said. If lipids are placed in a soup of biochemicals, they will encapsulate those materials when they self-assemble.
“This makes them a fantastic reaction vessel to execute complex biochemical reactions that ordinarily occur inside living bacteria,” he said.
They gave Danelon an ideal way to study cell division outside a living cell.
Division
In nature, cell division is a complex dance of proteins, the molecular machines that do the cell’s work. This starts with the Min system, a group of proteins that finds the cell’s midpoint so it divides evenly. The interaction among these proteins chases them to the cell’s poles, so the region with the fewest Min proteins is the cell’s middle.
One Min protein, MinC, keeps another protein, FtsZ, in check. But when the amount of MinC in the middle of the cell declines during division, FtsZ can attach to the middle of the cell membrane and form a Z-ring, which will eventually squeeze together and split the cell in two.
Some researchers have demonstrated these protein interactions by injecting purified proteins into vesicles. Danelon, however, had a more ambitious goal: to create a system that behaves more like a living cell. So, he encoded the genes for Min system proteins in a synthetic strand of DNA.
He then had his vesicles entrap the DNA, ribosomes (small structures that assemble proteins), and the dozens of proteins needed to produce Min system proteins. Once inside the vesicle, those proteins read the DNA, turn it into RNA instructions, and begin building the Min proteins with the ribosomes.
“The complexity of this approach is not in the DNA sequence, but in converting DNA into proteins,” Danelon said. “It involves more than 100 separate biochemical reactions.”
It took seven years of experiments to learn how to manage and sequence those reactions, but Danelon and his team, including Ph.D. students Elisa Godino and Jonás Noguera López, have now demonstrated a Min system in a synthetic vesicle. His lab is also close to forming a Z-ring as well.
“Now, we want to combine the two and have Min-assisted positioning of the Z-ring in order to have binary division,” Danelon said.
“This is the next challenge. Others in my lab are working on genes to synthesize lipids necessary for growth and proteins that replicate DNA. But it is not a cut-and-paste process. We really need to optimize conditions within the vesicle to have these different modules interface perfectly. This is where the next challenge lies.”
And we will see how many years it takes.