Scientists Use Algorithm to Peer Through Opaque Brains
(Originally published by The Rockefeller University)
June 26, 2017
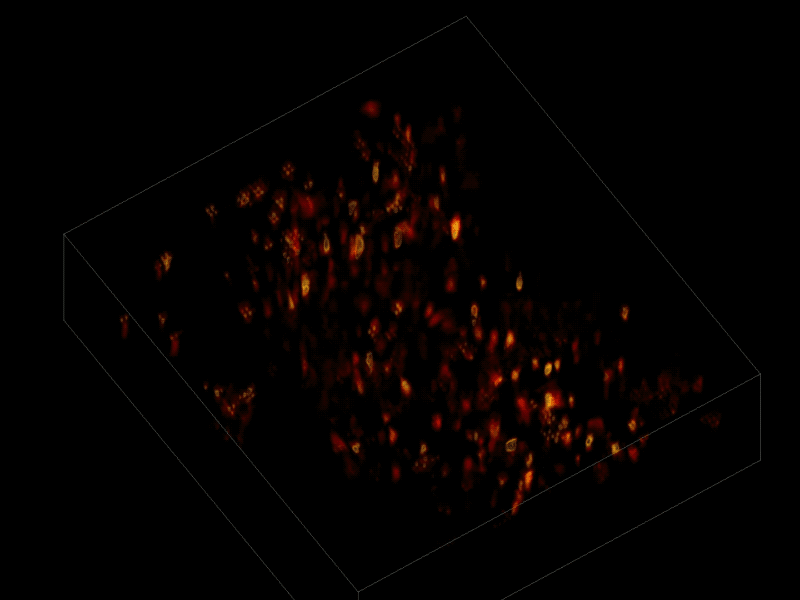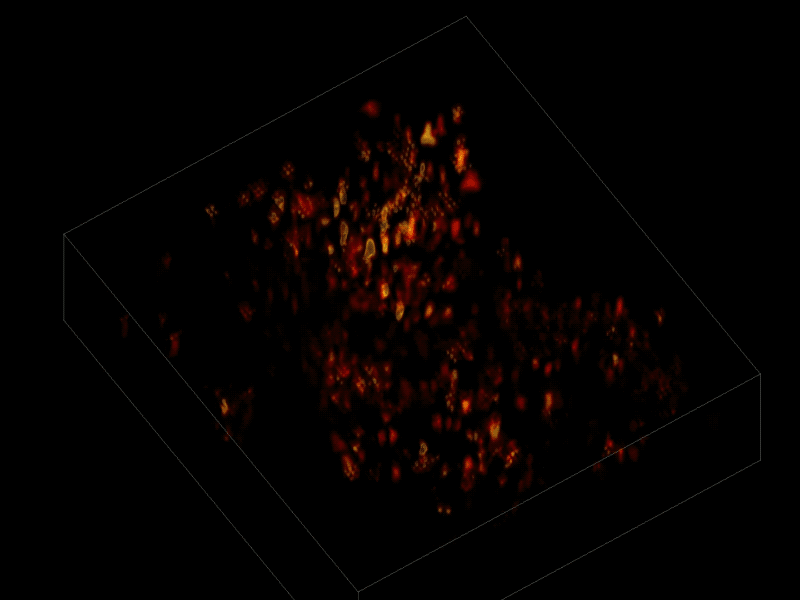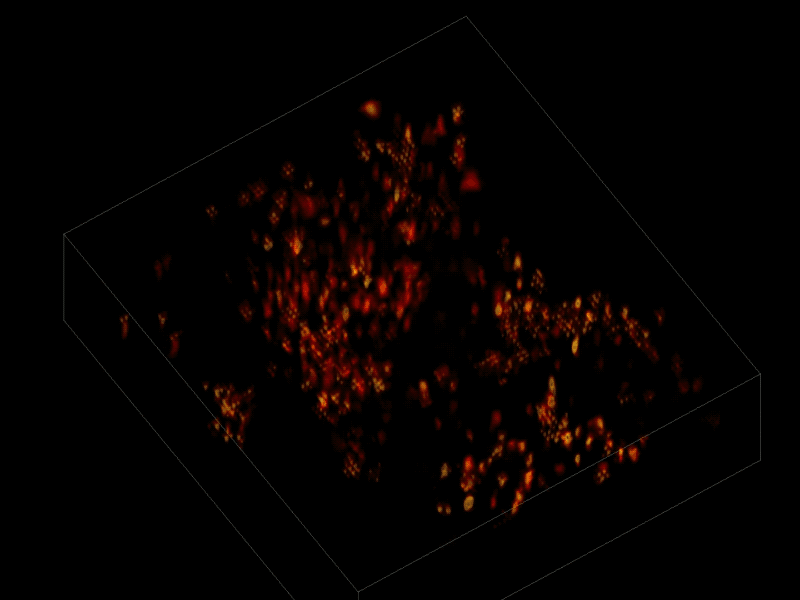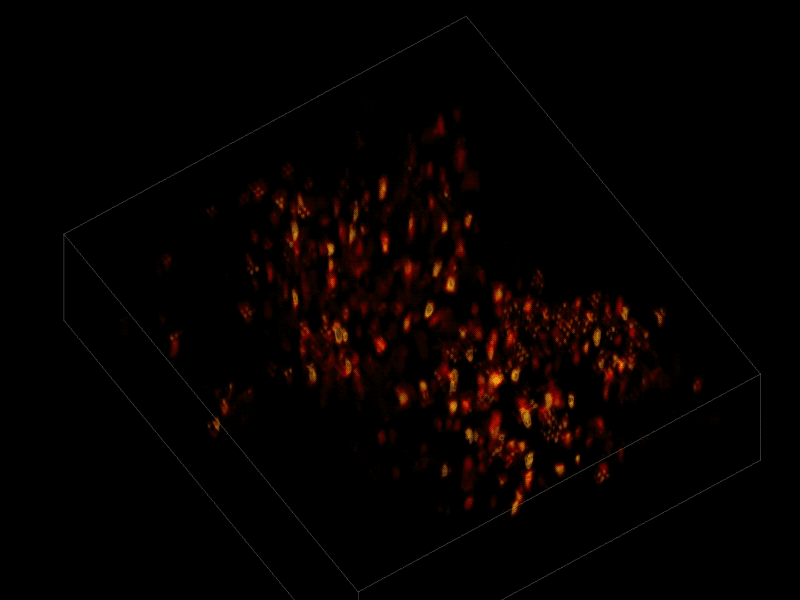
Trying to pinpoint signals from individual neurons within a block of brain tissue is like trying to count headlights in thick fog. A new algorithm, developed by researchers based at The Rockefeller University, brings this brain activity into focus.
In research described June 26 in Nature Methods, the team, led by Rockefeller’s Alipasha Vaziri, used a light microscope-based technique to capture neural activity within a volume of mouse brain tissue at unprecedented speed. The algorithm allowed them to pinpoint the signals from hundreds of individual neurons in a single recording.
“Our goal is to better understand brain function by monitoring the dynamics within densely interconnected, three-dimensional networks of neurons,” says Vaziri, head of the Laboratory of Neurotechnology and Biophysics and Associate Director of the Kavli Neural Systems Institute.
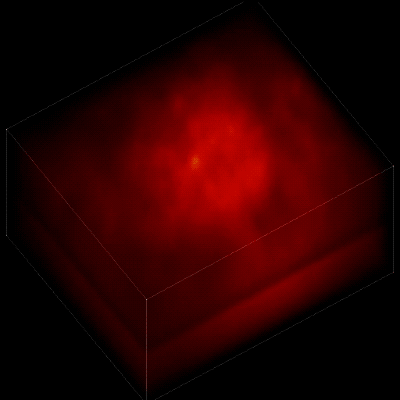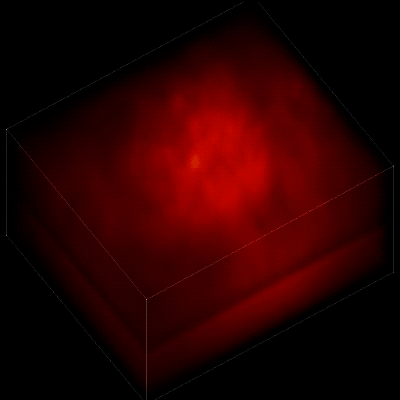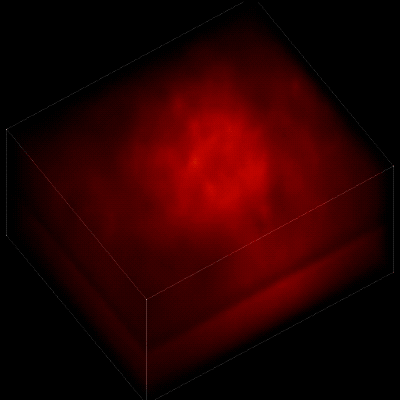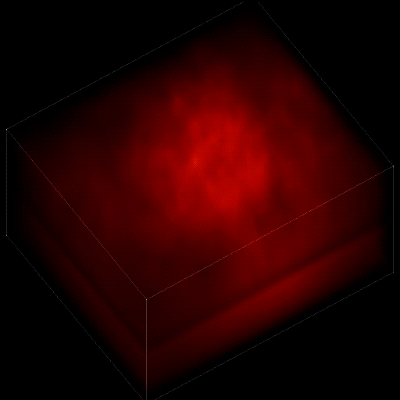
For this research, Vaziri and his colleagues engineered the animals’ neurons to fluoresce; the stronger the signal, the brighter the cells shine. To capture this activity, they used a technique known as light field microscopy, in which an array of lenses generates views from a variety of perspectives. These images are then combined to create a three-dimensional rendering.
When used to look down into a mouse brain, this method has a major challenge: To record the activity of all neurons at the same time, their images have to be captured on a camera simultaneously. But this makes it difficult to distinguish the signals emitted by all cells as the light from the mouse’s neurons bounces off the surrounding, opaque tissue. With an earlier version of this and other similar approaches, the neurons therefore typically show up as an indistinct, flickering mass—not much help to scientists who want to see how individual cells behave.
Vaziri and his colleagues devised a solution to this problem: a sophisticated algorithm that makes it possible to simultaneously capture both the location of the individual neurons and the timing of their signals.
In this way, Vaziri and his colleagues tracked the precise coordinates of hundreds of active neurons over an extended period in mice that were awake and had the option of walking on a customized treadmill. What’s more, they were able to capture signals within a three-dimensional section of brain containing multiple layers of neurons.
By dramatically reducing the time and computational resources required to generate such an image, the algorithm opens the door to more sophisticated experiments. “It may now be possible to alter stimuli in real time based on what we see going on in the animal’s brain,” Vaziri says.
Funding information:
This work was supported in part by the Vienna Science and Technology Fund Project, the Human Frontiers Science Program Project, the Institute of Molecular Pathology, The Kavli Foundation, and the Intelligence Advanced Research Projects Activity via Department of Interior/Interior Business Center.